EditR
estimating base editing by Sanger sequencing
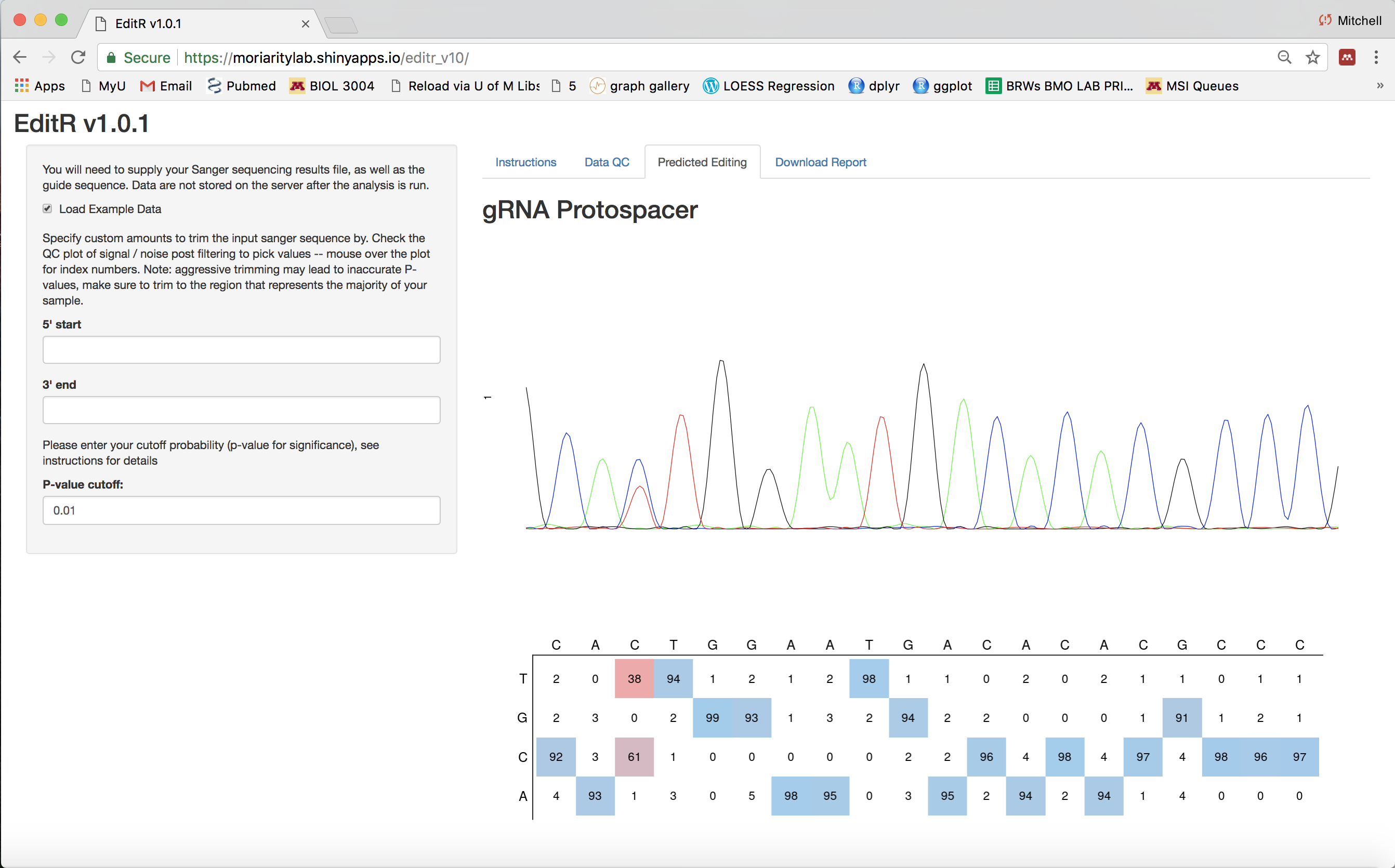
EditR
EditR is an algorithm for predicting potential editing in a guide RNA region from a single Sanger sequencing run. This allows users to estimate base editing efficiency quicker and cheaper than using deep sequencing.
It consists of the algorithm implemented in the R statistical programming language and provided as a Shiny app built to make this algorithm easy to use.
Citation
If this software helps you – please cite us and spread the word!
Kluesner M, Nedveck D, Lahr W, Moriarity B. EditR: A method to quantify base editing via Sanger sequencing. The CRISPR Journal. 2018.
Using the Shiny App
You can use the R Shiny App in two ways: through and instance on shinyapps.io, or installing it locally on your computer. We recommend testing it out on shinyapps.io, and then installing it locally to do a lot of analysis with.
Installing EditR locally
- Install R and R Studio (desktop version)
- Download code from github repository, and unzip into a directory you want to work from
- Open R Studio and run the code in dependencies.R to install the packages required
- In R Studio, open server.R, and click the run app button
- EditR should be started in your default web browser
Issues / problems
If there are issues with R, R Studio, installing R packages, or Shiny, please search for the error in your search engine of choice.
If you run into a problem with EditR, please feel free to submit and issue on GitHub, or contact us at baseeditr@gmail.com